




1) Molecular Modeling and Drug Designing
a) Homology Modeling:
It is the theoretical and computational approach to develop a sufficient accurate molecular three dimensional structure and analyzing their behavior at atomic level. It starts with the building of three dimensional structure using different calculation methods of computational chemistry. Homology modeling refers to creating 3-D structure of a protein using previously known homologous experimentally known structure. Multiple steps are involved in homology modeling like template identification, sequence alignment, backbone generation, ab-initio loop building, over all model optimization and model quality and validation.
b) Virtual Screening and Molecular Docking Simulations:
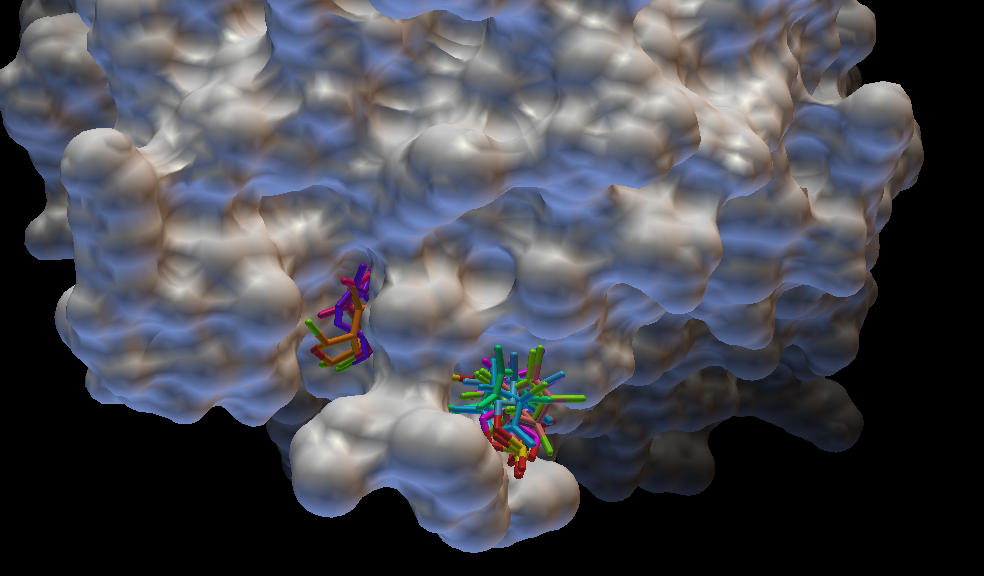
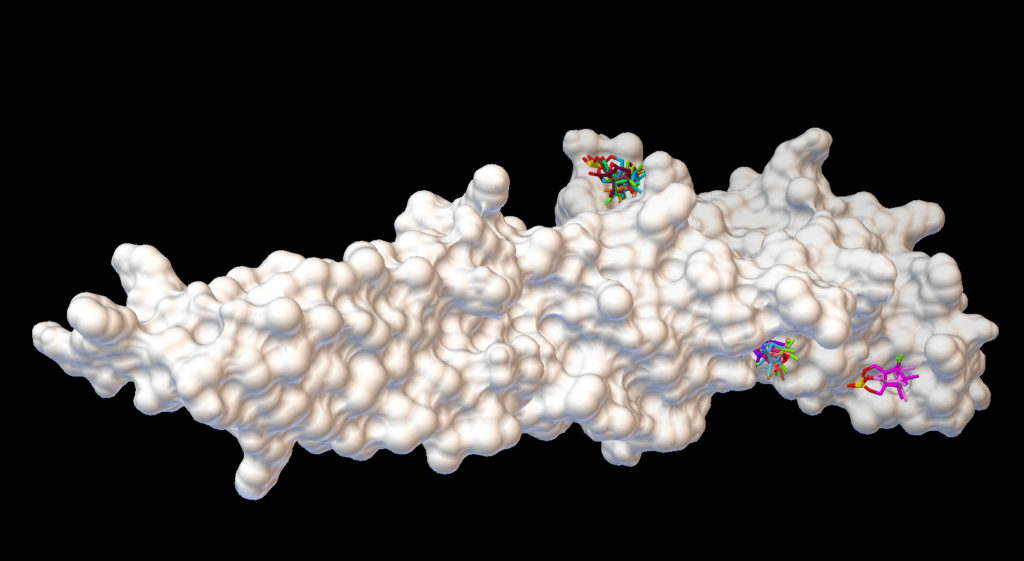
Molecular docking is known for predicting the best possible favorable orientation of binding of a drug or small peptide into the binding pocket of the target protein to form the most stable complex. Orientation of the ligand molecule in the protein target has a major role to play in determining its association strength, which is also known as binding affinity.Interactions between receptor and ligand are due to the forces like electrostatic forces, electrodynamics forces, static forces, solvent related forces. Virtual screening is a computational technique used in drug discovery research for screening of large chemical space for potential lead drug candidates towards a known drug target, usually an enzyme or a protein receptor. The appliance of virtual screening is being highly recognizing value in pharmaceutical industries due to its high accuracy and use of correlation method. Industries are growing towards screening of enormous chemical space of over 10 million chemical compounds. Based on the available knowledge regarding potential drug target/ligand, structure/ligand based virtual screening will be implemented either individually or in combination for identifying potential lead drug like compounds.
c) Free energy calculations:
Free energy is an important thermodynamic quantity to predict the difference between the energies of two states of a molecular system. Free energies constitutes the fundamental chemical quantities like binding constants, solubilities, partition coefficients and adsorption coefficients and it is expressed as the averages of ensembles of atomic configurations of molecular system of interest.
d) Protein-Protein interaction studies:
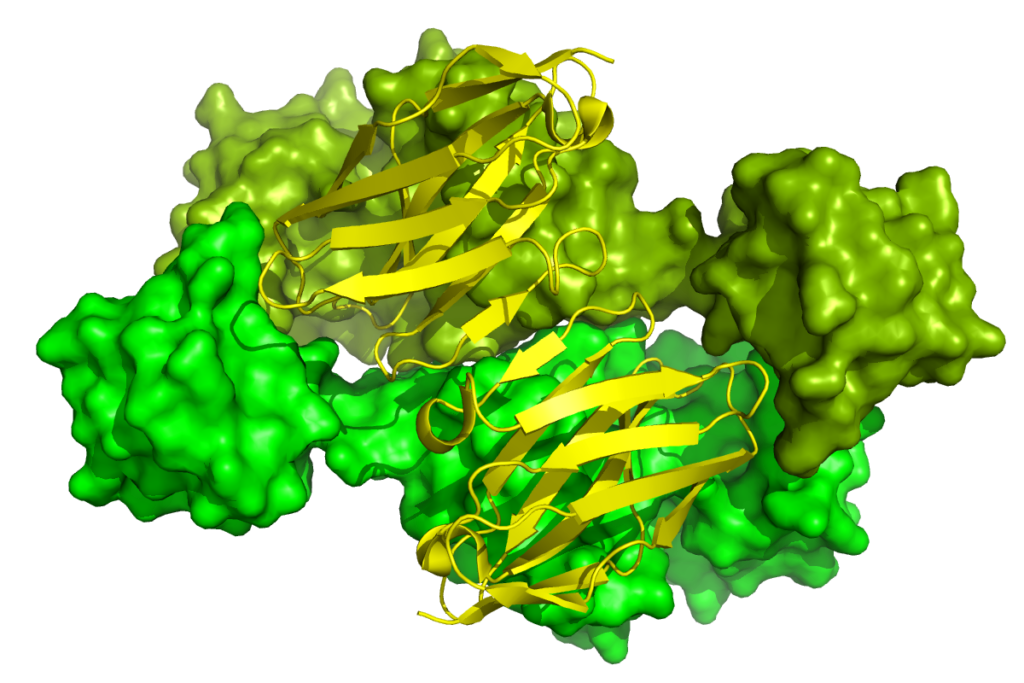
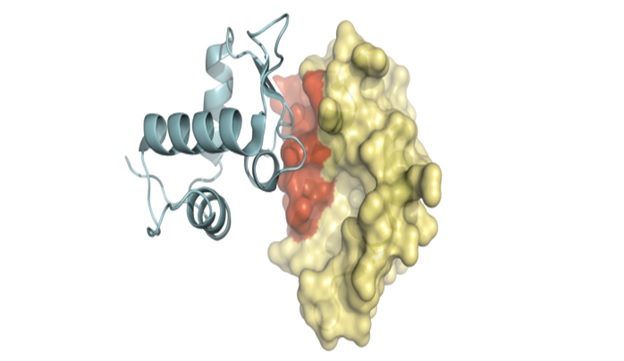
Protein-protein interaction plays very important role in determining the protein function and activity of target protein and drug ability of molecules. The interactomics of organism is the composition of numerousprotein-protein interactions and study of several interactions is significant measure in the direction of rational and ligand based drug designing. They can help us to illustrate protein complexes and pathways; interaction networks can be used as a draft ‘map’ to add detail to biological processes and pathways. Several properties of PPI such as allosteric sites and hotspots have been incorporated into drug-design strategies.
e) Molecular dynamic simulations
Molecular dynamic (MD) simulations is a method in which physical movements of various atoms and molecules under given physiochemical conditions like temperature and pressure, are allowed to interact and move for a period of time and their energy is calculated as perNewton’s law of motion. MD is generally used tocalculate the thermodynamic properties of the given protein structure in complex with/without ligand in the presence of solvent at given pH, temperature and pressure. Through this method total potential energy of the given protein or protein-ligand complex along with associated interaction energies can be calculated towards validating their association strength during the conformational changes.
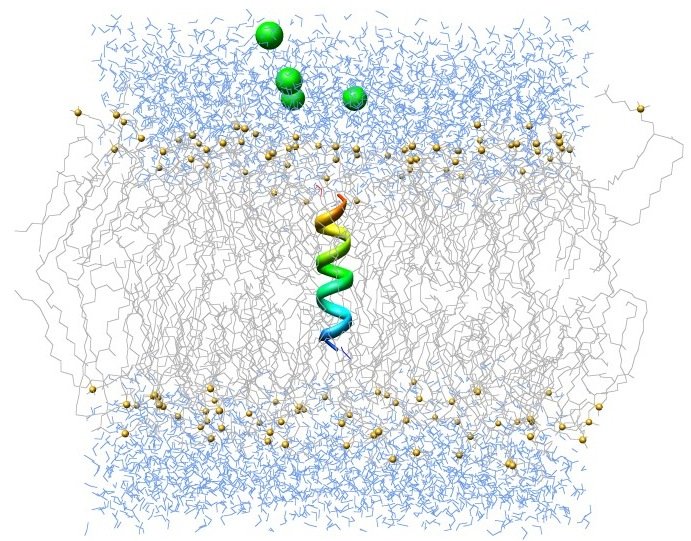
f) ADMET predictions
Accessing drug-likeness of a molecule is one of the crucial steps for optimizing pharmacokinetic and pharmaceutical properties of a lead molecule towards an active drug. Computational prediction of the ADMET (Absorption, Digestion, Metabolism, Excretion and Toxicity) properties is an efficient method to decrease the risk of chances of late stage abrasion and minimizing the safety problems of a drug. This is a cost effective method to assist drug discovery process and to remove molecules with poor ADMET properties from the drug development pipeline.
2) Molecular genomics
a) GWAS (Genome-wide Association Studies):
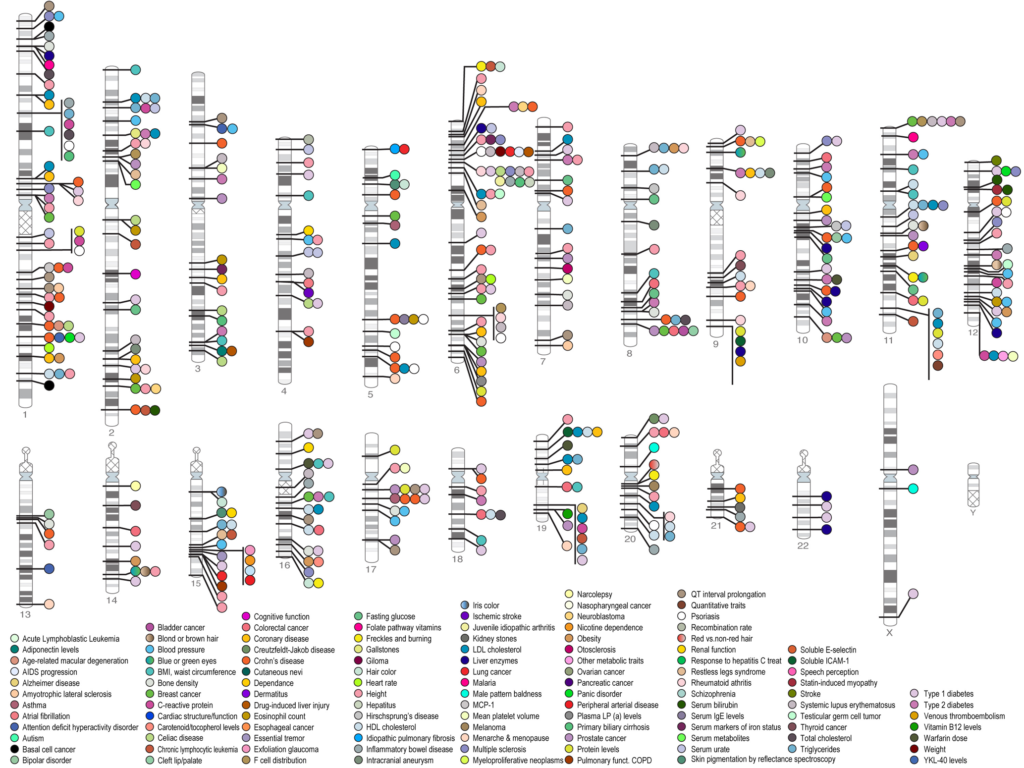
Genome-wide association studies includerecognition of the genes allied with the several human diseases or the genes that may be a factor for developing the risk of particular disease. In this tool DNA sequence variations are measured and analyzed from across the human genome and these small variations can be the SNP’s, failures of linkages for complex diseases etc. GWAS has been playing a successful role in the field of pharmacogenetics by identifying DNA sequence variations that are involved in the drug metabolism and efficacy.
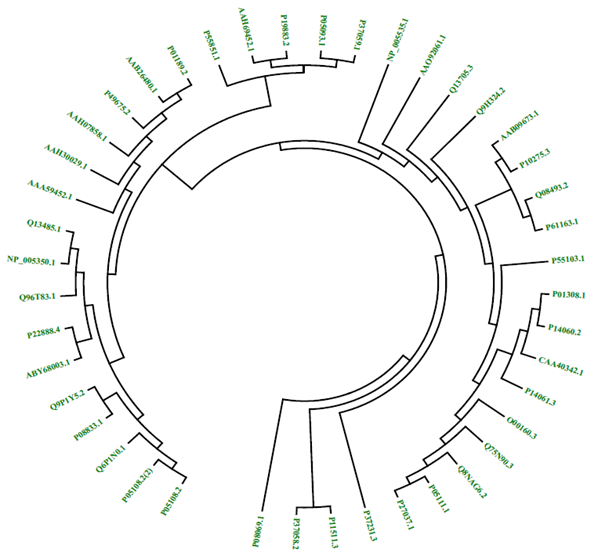
b) Phylogenomics:
Phylogenomics involves the reconstruction of evolutionary relationships by comparing sequences of whole genomes or portions of genomes. Basic fields that comes under phylogenomics are prediction of the gene function by generating phylogenetic trees, prediction and establishment of evolutionary relationships by constructing fully resolved phylogenetic trees and timing constraints can be recovered precisely, gene family evolution by comparing complete gene sets for a group of organisms and prediction andretracing lateral gene transfer.
c) Pharmacogenomics:
Pharmacogenomics embraces on the study of how genetic variations affect the person’s response to drugs also known as drug-gene testing. It deals with the genetic testing of the various genes and looking for the gene variants that the medicated drug could be an effective treatment for a person, looking for the side effects of the specific drug and its metabolism and toxicity. It also comprises improvement and personalization of the drug therapy.
d) Functional Genomics:
The functions and interactions of genes and proteins can be depicted by making use of genome-wide approaches i.e., functional genomics, in contrast to the gene-by-gene approach of classical molecular biology techniques. It focuses on the pattern of gene expression and interaction.Novel technologies that can come under functional genomics are interatomics, transcriptomics, proteomics etc.
3) Services: Scientific event management (conferences, seminars and workshops)
For any enquiry please contact us at: biomics.tech@gmail.com
